Stoichiometric network model of Corynebacterium glutamicum ATCC13032
last updated on 23.02.2015
In the 1950s, the Gram-positive rod-shaped bacterium Corynebacterium glutamicum was discovered in Japanese soil on the search for a natural L-glutamate producer. Since then, C. glutamicum turned into a versatile biotechnological working horse producing L-glutamate and L-lysine in multi-million-tons per year. Fast growth rates of up to 0.6 h-1 (Grünberger et al., 2013) qualified, the generally-regarded-as-save (GRAS) microorganism, C. glutamicum as a versatile amino acid producing workhorse for industrial biotechnology. Although C. glutamicum itself is non-pathogenic, it serves as a model organism for the sub-order Corynebacterianeae, which includes pathogenic species such as Corynebacterium diphtheriae, Mycobacterium tuberculosis and Mycobacterium leprae.
Intensive research heavily relying on traditional mutagenesis and selection strategies lead to a large collection of versatile production strains. The advent of DNA-sequencing technologies in 1977 marked a paradigm shift that unlocked targeted strain design by metabolic engineering. In 2003 the genome sequence of C. glutamicum became publicly available (Kalinowski et al., 2003). Nevertheless, it took until 2009 to reconstruct the first functional biochemical network models (Kjeldsen et al., 2009, Shinfuku et al., 2009). |
 |
The joint research consortia SysMap, FlexFit, and SysEnCor followed the systematic system biology approach to optimize C. glutamicum strains for the production of a broad range of amino acids as well as enhancing the strain's capabilities to utilize alternative carbon sources. As part of this research a consolidated and extended biochemical network model, termed iEZ475, has been built.
 |
||||
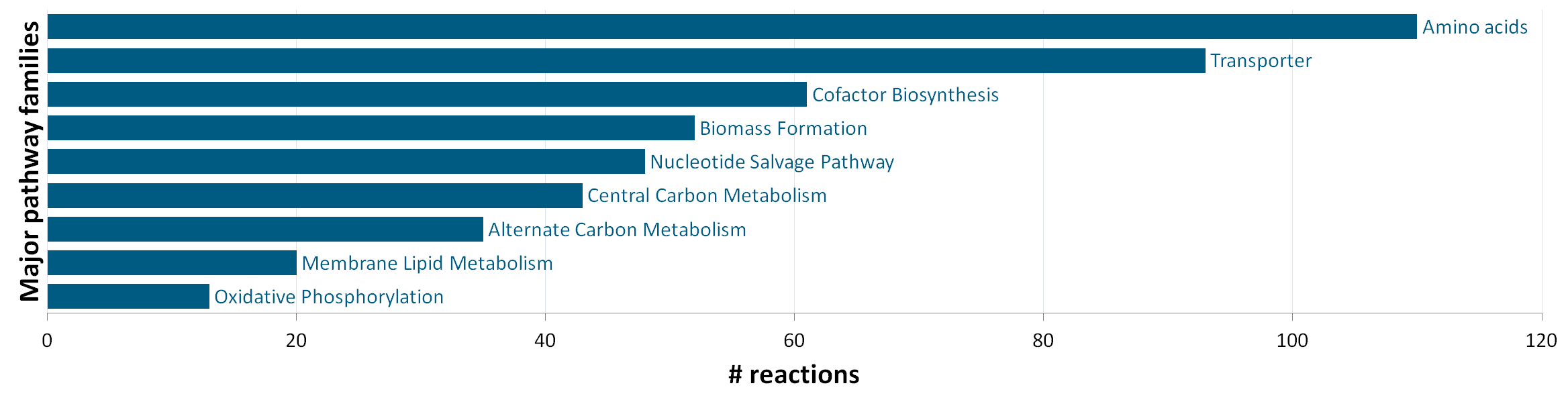
The stoichiometric model iEZ475 contains 475 metabolic reactions and 408 unique metabolites (340 intra- and 68 extracellular). The reactions are grouped in nine major pathway families: (1) central carbon metabolism, (2) amino acid synthesis, (3) oxidative phosphorylation, (4) membrane lipid metabolism, (5) nucleotide salvage pathway, (6) cofactor biosynthesis, (7) biomass formation, (8) alternate carbon metabolism (including the glycogen metabolism), and (9) transport reactions. The network model accounts for protein, DNA, RNA and cell-wall components as macromolecules, which are lumped in a final biomass equation based on their weight fraction of the biomass. |
The notation of the metabolites and reactions adopts the well-established E.coli-style (Feist et al., 2007). Importantly, in contrast to the published models (Kjeldsen et al., 2009, Shinfuku et al., 2009), the iEZ475 reactions are augmented with the balances for protons and water. All reactions, their cofactor-usage and transport steps have been carefully proofread. Summarizing, the current metabolic network iEZ475 represents a well-founded knowledge base to derive model-based predictions for C. glutamicum strain development.
Applying Flux Balance Analysis (FBA) on the network allows predicting metabolic flux distributions, growth rates, substrate uptake rates, and product secretion rates. One main value of the updated model is in its ability to predict phenotypes under a wider range of conditions than its predecessor. Thus, the model iEZ475 has been used to investigate capabilities, predict phenotypic behavior during different environmental and genetic conditions, and to test biological hypotheses. In particular, due to the added proton balances the validation focuses on energetic properties, i.e. cellular respiration and the phosphate/oxygen ratio (P/O ratio).
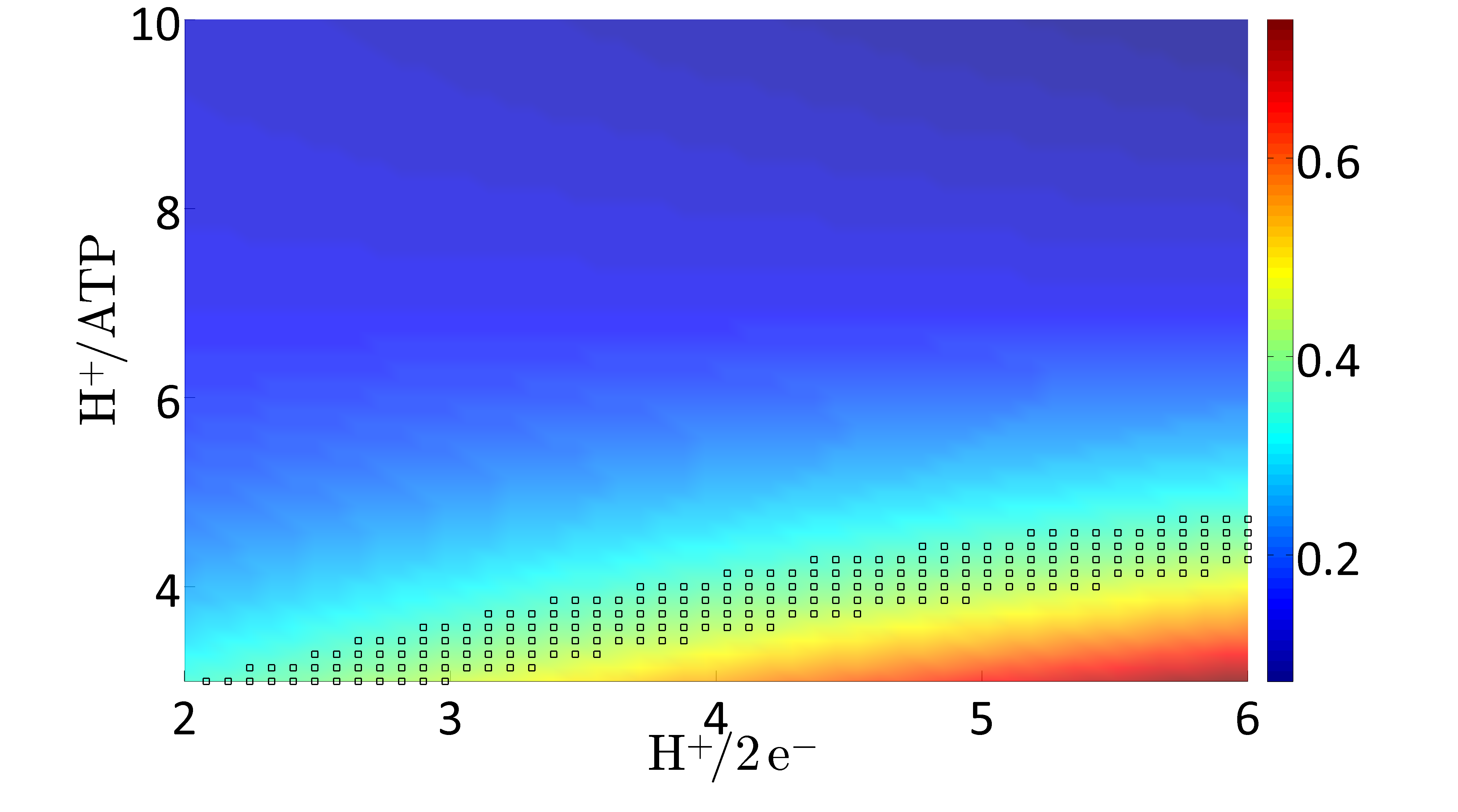 |
C. glutamicum exhibits a branched respiratory chain with two terminal oxidases. For investigating the impact of varying usage of the two terminal oxidases the H+/2 e- ratio is varied in the possible range between 2 (100% bd) and 6 (100% bc1-aa3). For each H+/2 e- ratio the amount of protons needed to synthesize one ATP (H+/ATP) is varied between 3 and 10 H+/ATP. For all value pairs of H+/2 e- and H+/ATP ratios the FBA problem is solved for the objective "maximize for growth". Here, the glucose uptake rate is fixed to an experimentally determined value of 5.4 mmol gCDW-1 h-1 (Koch-Koerfges et al., 2012). Model prediction of the optimal growth rate (μ[h-1] color coded) as function of the H+/ATP and the H+/2e- ratios, respectively, i.e., the ratios by which the two terminal oxidases are used. The marked area represents the experimentally measured growth rate μexp = 0.4 ± 0.01. |
References

- Bott, M., and Niebisch, A. (2003). The respiratory chain of Corynebacterium glutamicum. J. Biotechnol. 104, 129-153.
- Feist, A.M., Henry, C.S., Reed, J.L., Krummenacker, M., Joyce, A.R., Karp, P.D., Broadbelt, L.J., Hatzimanikatis, V.,
and Palsson, B.Ø. (2007). A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that
accounts for 1260 ORFs and thermodynamic information. Mol. Syst. Biol. 3, 121.
- Grünberger, A., van Ooyen, J., Paczia, N., Rohe, P., Schiendzielorz, G., Eggeling, L.,Wiechert, W.,
Kohlheyer, D., and Noack, S. (2013). Beyond growth rate 0.6: Corynebacterium glutamicum cultivated
in highly diluted environments. Biotechnol. Bioeng. 110, 220-228.
- Kalinowski, J., Bathe, B., Bartels, D., Bischoff, N., Bott, M., Burkovski, A., Dusch, N., Eggeling, L.,
Eikmanns, B.J., Gaigalat, L., et al. (2003). The complete Corynebacterium glutamicum ATCC 13032
genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J. Biotechnol. 104, 5-25.
- Kjeldsen, K.R., and Nielsen, J. (2009). In silico genome-scale reconstruction and validation of the
Corynebacterium glutamicum metabolic network. Biotechnol Bioeng 102, 583-597.
- Koch-Koerfges, A., Pfelzer, N., Platzen, L., Oldiges, M., and Bott, M. (2013). Conversion of Corynebacterium glutamicum
from an aerobic respiring to an aerobic fermenting bacterium by inactivation of the respiratory chain. Biochim. Biophys. Acta 1827, 699-708.
- Shinfuku, Y., Sorpitiporn, N., Sono, M., Furusawa, C., Hirasawa, T., and Shimizu, H. (2009). Development
and experimental verification of a genome-scale metabolic model for Corynebacterium glutamicum. Microb. Cell Fact. 8, 43.
- Zelle, E., Nöh, K., Wiechert, W. (2015) Growth and Production Capabilities of Corynebacterium glutamicum: Interrogating a Genome-scale Metabolic Network Model. In A. Burkowski (Eds.), Corynebacterium glutamicum: From Systems Biology to Biotechnological Applications (39-54). Horizon Press
Download Area
Before download we would kindly ask for name and email address. If you would like to be informed about future updates please enter affiliation, too.
icons by famfamfam.com last change: 29.09.2023 | IBT Webmaster | Print


